This should be correspond to every second file. It uses the mapping results from Bowtie to identify splice junctions between exons.

Rna Seq Course Alignment Using Tophat Old Youtube
This is quite different conceptually to mapping to the transcriptome directly.

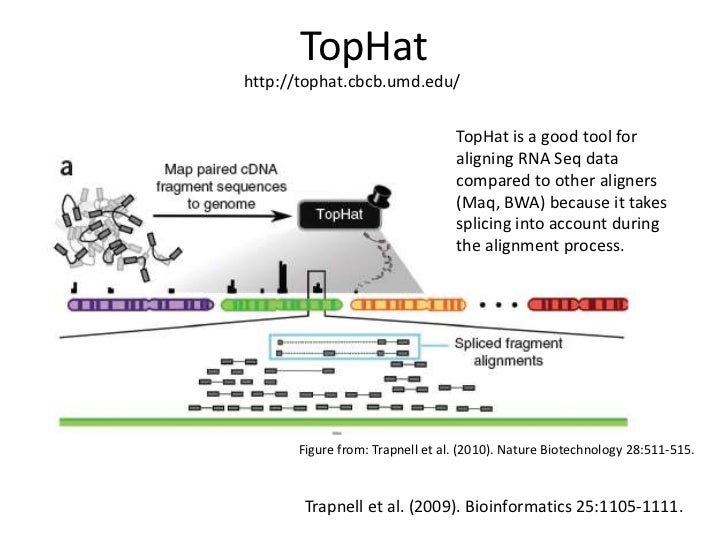
. TopHat2 uses using Bowtie to align RNA-Seq reads to mammalian-sized genomes and then analyzes the mapping results to identify splice junctions between exons. The requirements for aligning this type of data is slightly different from eg. If theres no index for your organism its easy to build one yourself.
One of CBSU BioHPC Lab workstations has been allocated for your workshop exercise. In this post we will align the paired end data to the human genome with Tophat. It is slow but consumes less memory.
Understand QC steps that can be performed on RNA-seq reads. Tophatcmdwithmetadatapastetophat -G gtf -p 5 -o outputdir libraryName bowidx fastqDirfastqnnnnsep sinkfiletophat-commandsshtypeoutput catbinsh nnnn cattophatcmd sink. Using TophatCufflinks to analyze RNAseq data.
This practical will introduce some popular tools for basic processing of RNA-seq data. Click on the multiple datasets icon and select all six of the forward FASTQ files ending in 1fastq. Generate interactive reports to summarise QC information with MultiQC.
This tutorial from 2017 covers the TopHat aligner. Press the key to enter command mode. The allocations are listed on the workshop exercise web page.
Quick Start Install the plugin by downloading the gplugin file and dragging it in to Geneious or use the plugin manager in Geneious under Tools - Plugins in the menu. Data analysis step 3. TopHat is a collaborative effort between the University of Maryland Center for Bioinformatics and.
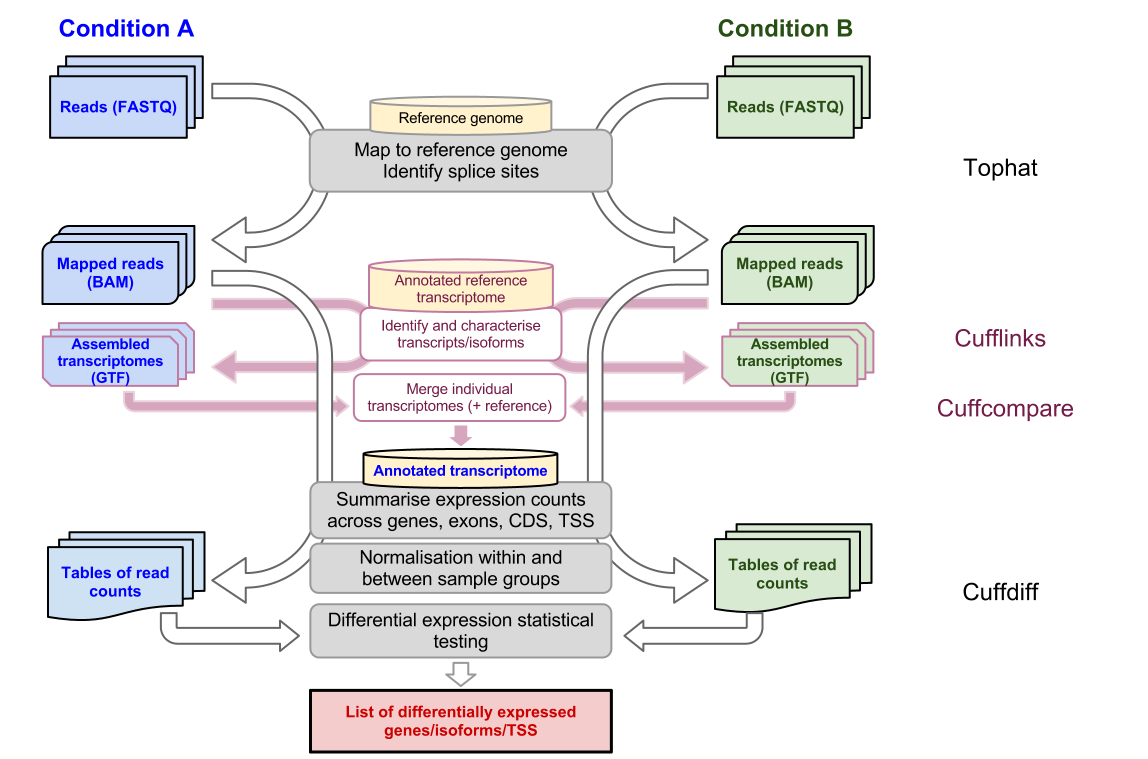
RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Hands-on using TopHat and Cufflinks in Galaxy Tutorial 3 Advanced RNA-Seq Analysis topics. A newer more advanced worfklow was introduce with Cufflinks version 220 and is shown on the right. We recommend that you watch the video Aligning RNA-seq reads to reference genome instead which covers t.
The following script creates the TopHat commands necessary for the alignments. Background Web Resources. It aligns RNA-Seq reads to mammalian-sized genomes using the ultra high-throughput short read aligner Bowtie and then analyzes the mapping results to identify splice junctions between exons.
In the left tool panel menu under NGS Analysis select NGS. Both are still supported. Filtering raw alignments step 1 Tophat produced an bam file at output_directoryaccepted_hitsbam.
Press the i key to enter insert mode. Go to an empty line with you cursor and copy paste the new RNA_HOME and PATH commands into the file. Tophat -o output_directory -p1 genome_database rnaseqfastq.
The Bowtie site provides pre-built indices for human mouse fruit fly and others. TopHat is a fast splice junction mapper for RNA-Seq reads. Use the Galaxy Rule-based Uploader to import FASTQs from URLs.
You first need to build an index file for your genome. This tutorial will focus on doing a 2 condition 1 replicate transcriptome analysis in mouse. If you would like to learn more about how to use vi try this tutorialgame.
Afterwards align the RNAseq data to the genome. Align the RNA-seq short reads to a reference genome. If you have Bowtie 2 installed and want to use it with Tophat v20 or later you must create Bowtie 2 indexes for your.
To find junctions with TopHat youll first need to install a Bowtie index for the organism in your RNA-Seq experiment. There are several types of RNA-Seq. Httpcbsutccornelleduww1sessionaspxwid9sid12 Please consult the PDF file with instructions on how to access and use the Lab workstations for the.
These lectures also cover UNIXLinux commands and some programming elements of R a popular freely available statistical software. A set of lectures in the Deep Sequencing Data Processing and Analysis module will cover the basic steps and popular pipelines to analyze RNA-seq and ChIP-seq data going from the raw data to gene lists to figures. Inspect the files in the working directory workdirmy_user_ID.
STAR is much faster but need a machine with large memory 30GB for human genome. Make use of Galaxy Collections for a tidy analysis. If you are not in working directory already type cd workdirusre_user_ID first ls.
Some of the applications used in RNA sequencing analysis are the following. Is this single-end or paired-end data. TopHat is a fast splice junction mapper for RNA-Seq reads.
Tophat is a splicing aware aligner so we can map transcripts to the genome. The left side illustrates the classic RNA-Seq workflow which includes read mapping with TopHat assembly with Cufflinks and visualization and exploration of results with CummeRbund. In this series of posts were going through an RNA-seq analysis workflow.
Much of Galaxy-related features described in this section have been developed by. Create a Galaxy Workflow that converts RNA-seq reads into counts. At the very end we can compare these results to the results we got from mapping directly to the.
More information on Tophat can be found here. Align paired end RNA-seq with Tophat. Transcriptome splice-variantTSSUTR analysis microRNA-Seq etc.
Jeremy Goecks Galaxy RNAseq tutorial httpmaing2bxpsueduujeremypgalaxy-rna-seq-analysis-exercise. So far weve downloaded and inspected sequence quality. RNA Analysis Tophat and set the parameters as follows.
Type wq to save and quit vi. Bowtie1 is an ultrafast memory-effi cient aligner for large sets of short reads. Tophat is a splice-aware mapper for RNA-seq reads that is based on Bowtie.
Part 1 is to get a suitable reference genome sequence. TopHat is a collaborative effort among Daehwan Kim and Steven Salzberg in the Center for Computational Biology at. It aligns RNA-Seq reads to mammalian-sized genomes using the ultra high-throughput short read aligner Bowtie and then analyzes the mapping results to identify splice junctions between exons.
RNA-seq transcriptome sequencing is a very powerful method for transcriptomic studies that enables quantification of transcript levels as well as discovery of novel transcripts and transcript isoforms. TOPHAT is widely used in the early days of RNA-seq data analysis. We rst cover a full work ow from reads.
Once installed run the plugin by selecting your reads and reference sequence then clicking on AlignAssemble - Map to Reference in the toolbar. RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Advanced RNA-Seq Analysis topics Hands-on tutorials Analyzing human and potato RNA-Seq data using Tophat and Cufflinks in Galaxy. In this tutorial well map reads from an RNA-seq study in Drosophila melanogaster to the reference genome using tophat.
This tutorial is inspired by an exceptional RNA seq course at the Weill Cornell Medical College compiled by Friederike Dündar Luce Skrabanek and Paul Zumbo and by tutorials produced by Björn Grüning bgruening for Freiburg Galaxy instance. Press the esc key to exit insert mode. Paired-end as individual datasets RNA-Seq FASTQ file forward reads.
Basic Analyses With Tophat Cufflinks Rnaseq Tutorial 1 Documentation

Reference Based Rnaseq Data Analysis Long
Imgc2011 Bioinformatics Tutorial
Aligning Rna Seq Data Ngs Analysis
The Cufflinks Rna Seq Workflow
Introduction To Bulk Rnaseq Analysis Bioinformatics Documentation
Incorporating Rnaseq Tophat To Augustus Computational Biology
0 comments
Post a Comment